
Wow, I haven’t written a blog in ages! Recently, Guildford had a snow day and it was such a surprise to wake up and look out my window to see snow everywhere. You best bet I was out to take some pictures because a proper snow day in the South East is quite rare! Regardless, I had my mask on of course.

Putting away the snow day, I’ve only just finished all of my exams for 2020/21 Semester 1, and boy am I glad to be free (but not really)! The minute I finished my final exam, I immediately took down all the relevant post-it notes I had pasted on the wall. It kind of breaks my heart to do that though; my wall feels a little too empty now. Anyway, the short break after exams gives me time to work on my research project before starting Semester 2.
Just for your information, I am in my final year of BSc (Hons) Biomedical Science and we are required to do a research project/dissertation in our final year. It is an academic year-long project and it is equal to two module’s worth of grades. Because of that, we only have three modules per semester in final year, as opposed to the normal four modules in previous years (or other courses).
My Research Project!
Onto my research project, it is actually pretty relevant to what is going on today! For my fellow science peers, my project is essentially an in silico (meaning “by computer modelling/stimulation”) investigation on whether certain enzyme inhibitors would work to treat SARS-CoV-2 (COVID-19). For those who aren’t familiar with the terms, I am basically using computer simulations to test whether certain pharmaceutical drugs would be able to bind to and block the action of the one of the virus’s proteins that facilitates entry into our cells.
Due to my project being an in silico/computational investigation, I don’t need to use a laboratory to collect the data I need. There are definitely other project title choices that do involve laboratory work, so worry not! When the project titles are revealed for you to choose in final year, they include tags such as “laboratory work”, “computational”, or “field work”, so narrowing down project title choices is made much easier. “Laboratory work” speaks for itself, but what about “field work”? Well, it generally involves travelling or visiting somewhere for you to collect the information and data for your project. For example, a few title choices required you to travel to a hospital to use the NHS computers to collect data from electronic patient records, alongside interacting with the NHS staff for additional data.
Some time after submitting your choices in Semester 1, your project title will be assigned to you. Your project supervisor will probably contact you to arrange a first meeting together with any other students that were assigned the same title, if there are any. The supervisor will brief you on the project and what your timeline for the project should be. In my case, I was told that I should have my introduction and methods section done by the end of January (which is about now, at the time of writing this), and the results section should be done by the time Easter break starts, leaving me the break to finish the project up. It is your initiative in setting up meetings with your project supervisor however, and this is particularly important because your supervisor’s assessment of you is a part of the grade rubric. It takes up 20% of the grade, so it is definitely a big portion!
The Boring Interesting Work on My Project
During the winter break, I worked on collecting data with certain computer programmes. Since my work involves binding between molecules, I had to learn to use AutoDock Vina as well as Discovery Studio Visualizer. AutoDock Vina is an open-source program for molecular docking where you input the two molecules you want to investigate and the program will determine whether the two will bind together. To do this however, I need to use Discovery Studio Visualizer to model the molecules I am studying to be able to input it into AutoDock Vina.
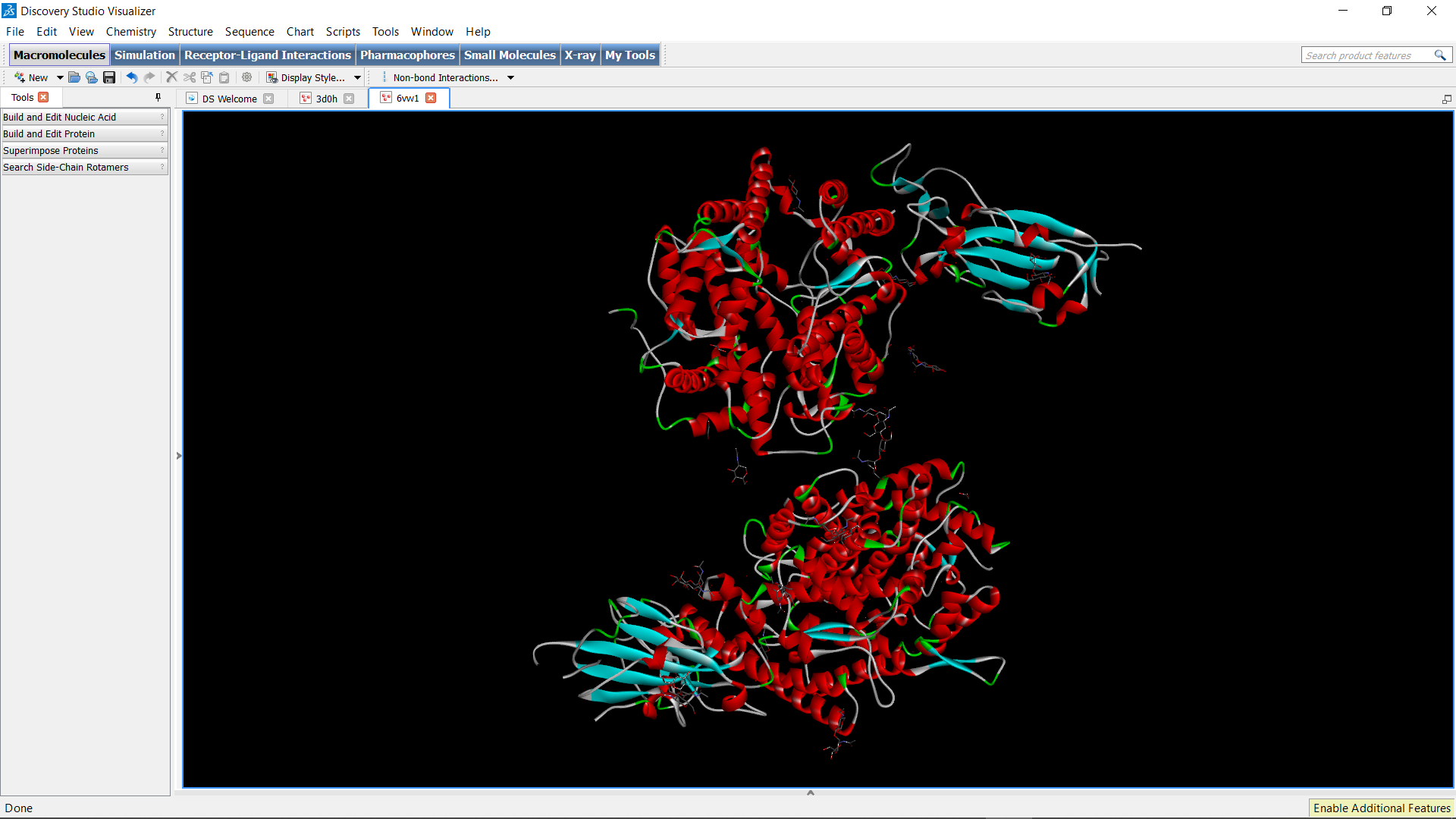
So, let’s say I am studying the ACE2 cellular receptor which facilitates the entry of SARS-CoV-2 (COVID-19) into the cell. I must first search for the exact structure of this particular receptor on a database called Protein Data Bank (PDB), where I can then download the file containing its crystal structure and input it into Discovery Studio Visualizer. From there, I can select and find out the parts of the molecule, as well as edit any molecules within the protein (like deleting any water molecules, etc.).
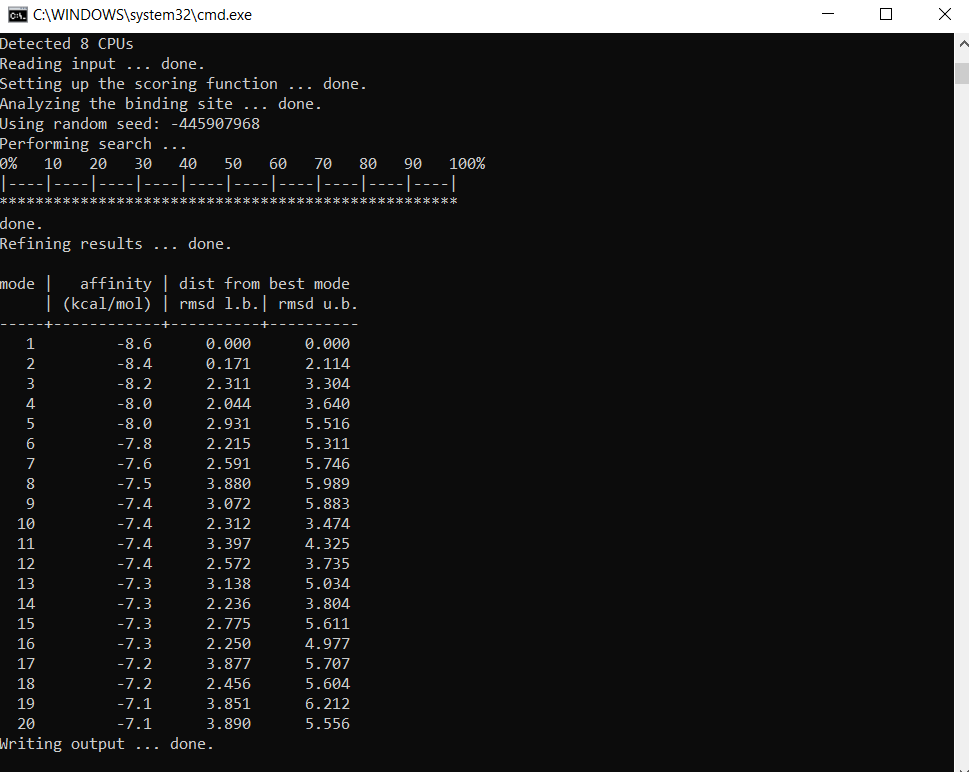
After all that editing, I run AutoDock Vina from command prompt and see what results I get! Basically, the program tries different binding conformations, and produces the ‘affinity (kcal/mol)’ column which is the strength of binding for that particular binding conformation.
That’s essentially the basics of the computational work that I have to do for my project! I really hope this wasn’t too much of a mind-boggle but don’t worry, I had to take some time to wrap my head around it at first too. I will probably start writing more again after this so keep a look out for any other updates!

Hi! I’ve found this blog interesting and I would like to know more about it. I’ve tried to send an email to malaysia@surrey.ac.uk but it was shown that it this is a n incorrect email. Could you please send me an email or provide me the correct one? Thank you.
Hi there, I sincerely apologise for that as there seems to be some technical issues with our mailbox. We will try to get it fixed as soon as possible, but I am able to answer any burning questions here if you’d like!